Customizing user environments
It is possible to customize the notebook environment using either a conda/mamba environment, or preamble scripts that execute before your notebook starts.
Preamble scripts
You can customize your EAF Jupyter installation with your own custom preamble scripts. These scripts are executed when you launch a notebook with the standard python (ipykernel) kernel. We provide a duplicate of the standard python kernel, labelled safemode, that ignores the preamble scripts on launch.
Why would I need this?
It may be useful to set up your environment via setup scripts from CVMFS. For example, you may want to execute a script like this:
source /cvmfs/dune.opensciencegrid.org/products/dune/setup_dune.sh
setup dunesw v09_52_00d00 -q e20:prof
Usage
The ipykernel launcher will execute two different preamble scripts in order:
~/.preamble/global.sh, which is executed on every notebook flavor.
~/.preamble/${NB_PROFILE}.sh
The second script allows you to customize notebook flavor-dependent scripts, based on the value of
the NB_PROFILE environment variable.
Note
The preamble scripts only run for the “python-ipykernel” launcher. See conda preamble scripts for information on running preamble scripts in a conda/mamba environment.
Example
~/.preamble/global.sh:
export FOO=globally-set
export BAR=globally-set
~/.preamble/astro-sl7-unpriv-interactive.sh:
export BAR=locally-set
The second preamble will only run for an astro notebook server; BAR is overwritten:
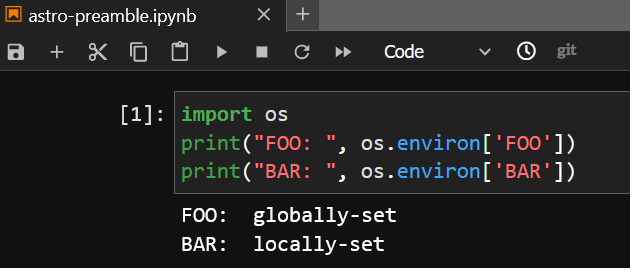
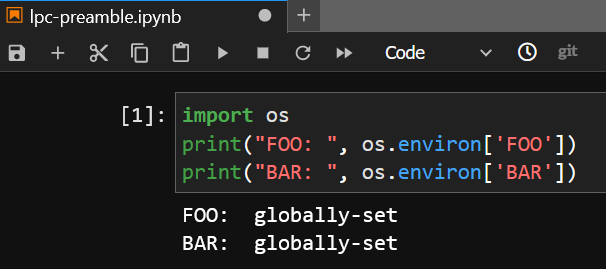
If you execute from an LPC notebook server, the second script does not execute and BAR
remains unchanged:
Conda/mamba
It is possible to customize your environment using anaconda and access that environment in your notebooks.
Configuring conda/mamba (only needed once)
From a terminal launcher, run mamba init. Then, write to .bash_profile in your home directory with
the following content:
# Get the aliases and functions
if [ -f ~/.bashrc ]; then
. ~/.bashrc
fi
Using conda/mamba
Conda is well-documented at https://conda.io (its use is outside the scope of this documentation). You can safely
substitute mamba for conda wherever it is listed; it is a faster reimplementation in C++ of conda.
Installing launchers
You must install the ipykernel package in order to make your custom environment available as a notebook kernel. After a minute or so, a new launcher should appear corresponding to your environment; it may be necessary to reload the web page.
Customizing conda with user scripts
For conda environments, you can add preamble scripts to ${CONDA_PREFIX}/etc/conda/activate.d; these will run before
your notebook starts. For example, in the snowflakes conda environment example below,
additional preamble scripts could be added to ~/.conda/env/snowflakes/etc/conda/activate.d.
Example: installing biopython in the snowflakes conda environment
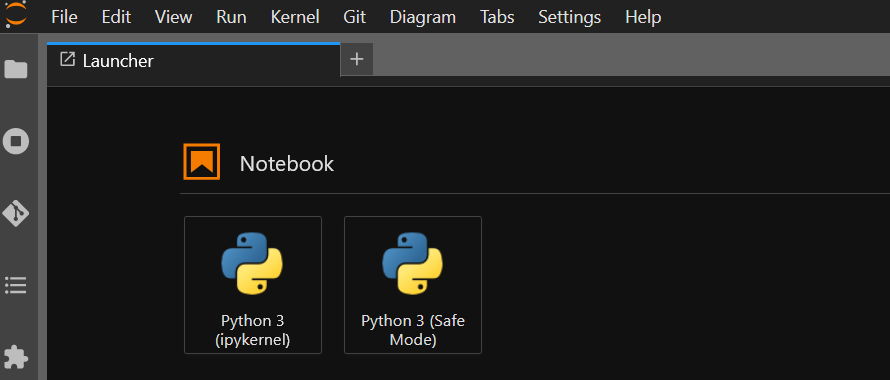
The vanilla launcher screen shows the two default launchers: python, and python-safemode.
We then install a new environment named snowflakes, including the biopython package:
[burt@jupyter-burt-lpc ~]$ mamba create -q -y -n snowflakes biopython ipykernel
Package Version Build Channel Size
────────────────────────────────────────────────────────────────────────────────────────────────────
Install:
────────────────────────────────────────────────────────────────────────────────────────────────────
+ _libgcc_mutex 0.1 conda_forge conda-forge/linux-64 Cached
+ _openmp_mutex 4.5 2_gnu conda-forge/linux-64 Cached
[ ... snipped for brevity ... ]
+ zeromq 4.3.4 h9c3ff4c_1 conda-forge/linux-64 Cached
Summary:
Install: 66 packages
Total download: 3MB
────────────────────────────────────────────────────────────────────────────────────────────────────
Preparing transaction: ...working... done
Verifying transaction: ...working... done
Executing transaction: ...working... done
The newer launcher now appears, with its full name visible via tooltip, indicating the name of the conda environment:
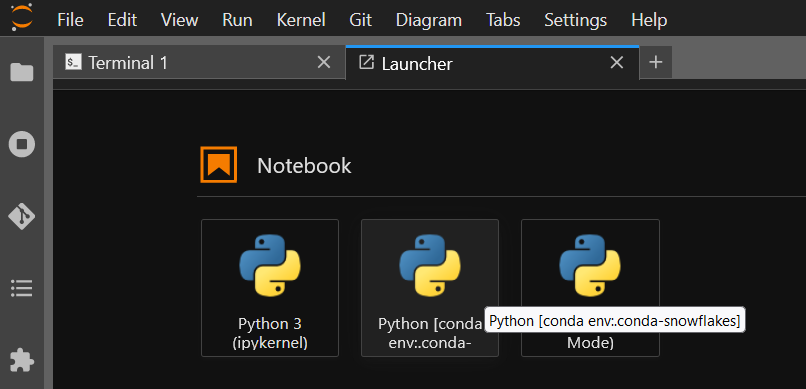
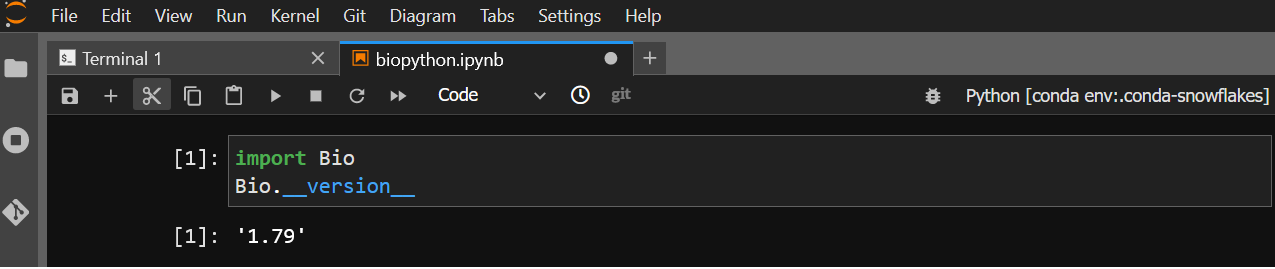
And the Bio module from biopython is available:
Pip
It is possible to customize your environment using pip install --user, but there are some caveats.
Warning
In order to allow user installation of pip packages, the system creates
~/.bash_profile and ~/.ipython/profile_default/startup/00-python-user-site.py files at first login.
If these files are removed or the automatically-generated content is modified, pip may no longer function properly. To have the
system regenerate the appropriate content, remove the ~/.python_no_user_site_unset file and launch a new server.
You may wish to set PYTHONUSERBASE to something other than $HOME to avoid conflicting packages from
being installed when working in different environments. For example:
[burt@jupyter ~]$ cd ~/work1
[burt@jupyter work1]$ PYTHONUSERBASE=$PWD/.local pip -q install --user numpy==1.24.1
[burt@jupyter work1]$ cd ~/work2
[burt@jupyter work2]$ PYTHONUSERBASE=$PWD/.local pip -q install --user numpy==1.21
To work in the work2 environment with NumPy 1.21, one can add the following notebook cell:
pythonuserbase = "/home/burt/work2/.local"
import sys
pyver = f"python{sys.version_info.major}.{sys.version_info.minor}"
sys.path.insert(0, f"{pythonuserbase}/lib/{pyver}/site-packages")